Mette Soendergaard • June 24, 2022
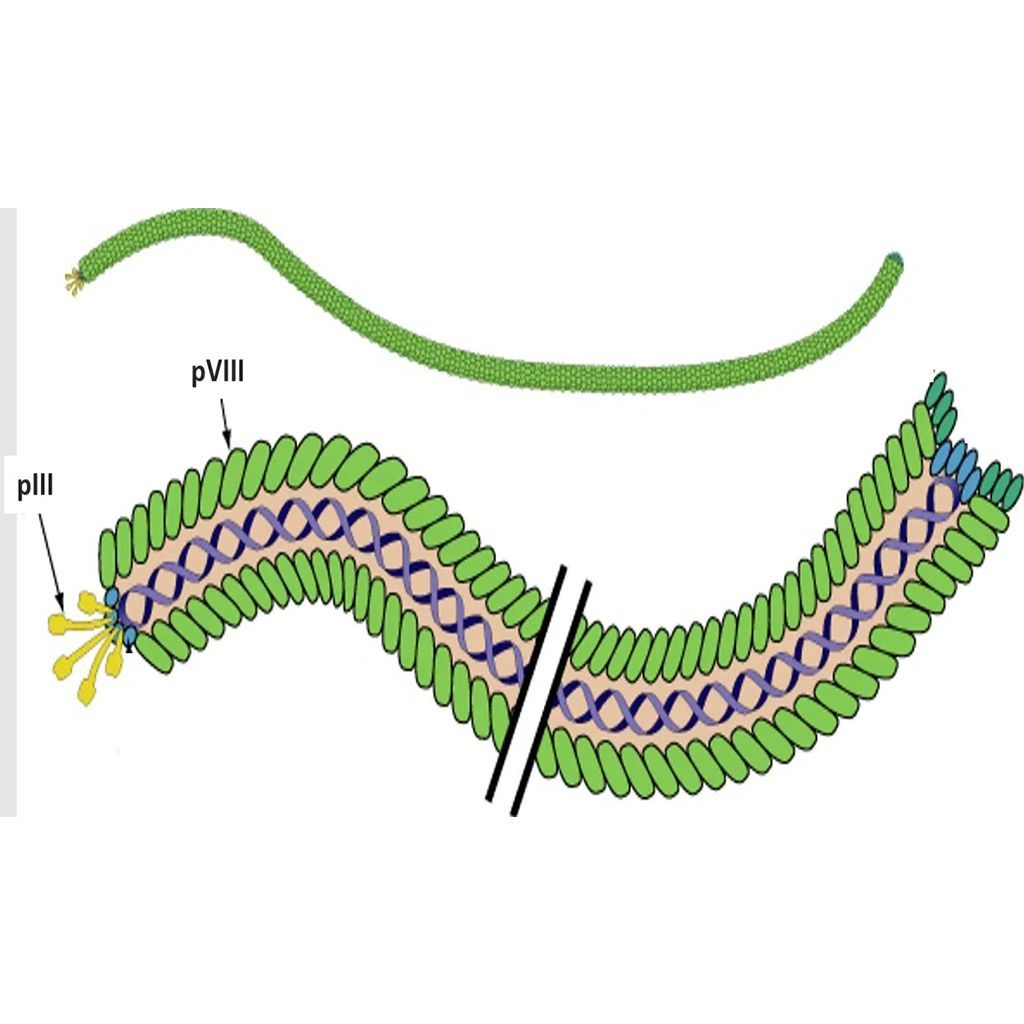
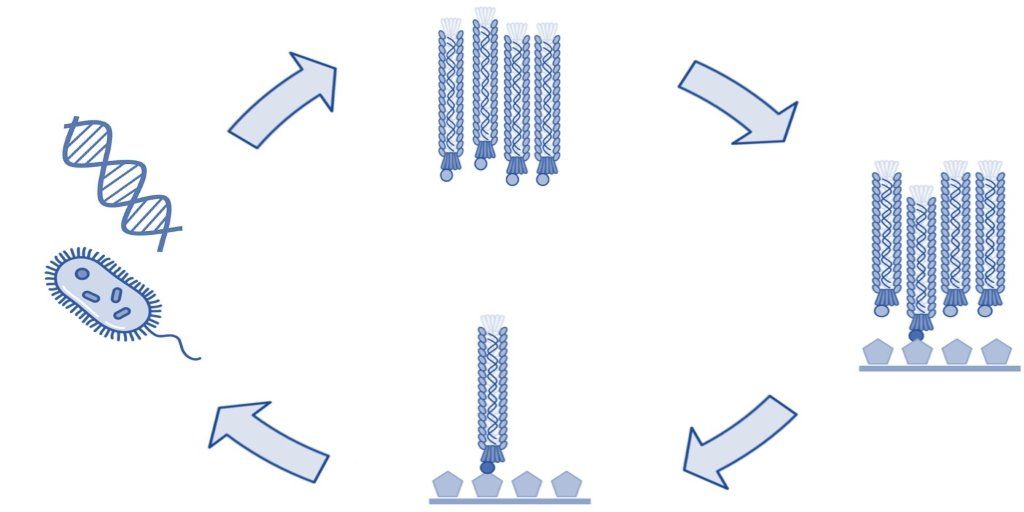
Filamentous Phage
In 1985, Dr. George Smith (University of Missouri) created phage display technology by genetically modifying the filamentous phages of the Ff class [1]. He worked specifically with the Fd species, which he had years earlier modified to create the Fd-tet cloning vector [2]. In addition to the Fd species, the Ff class also includes the filamentous M13 bacteriophage that has since been used extensively in phage display technology [3,4]. Both the bacteriophages M13 and Fd have been successfully used to assemble protein libraries to display peptides and antibodies and utilized to select high affinity ligands.
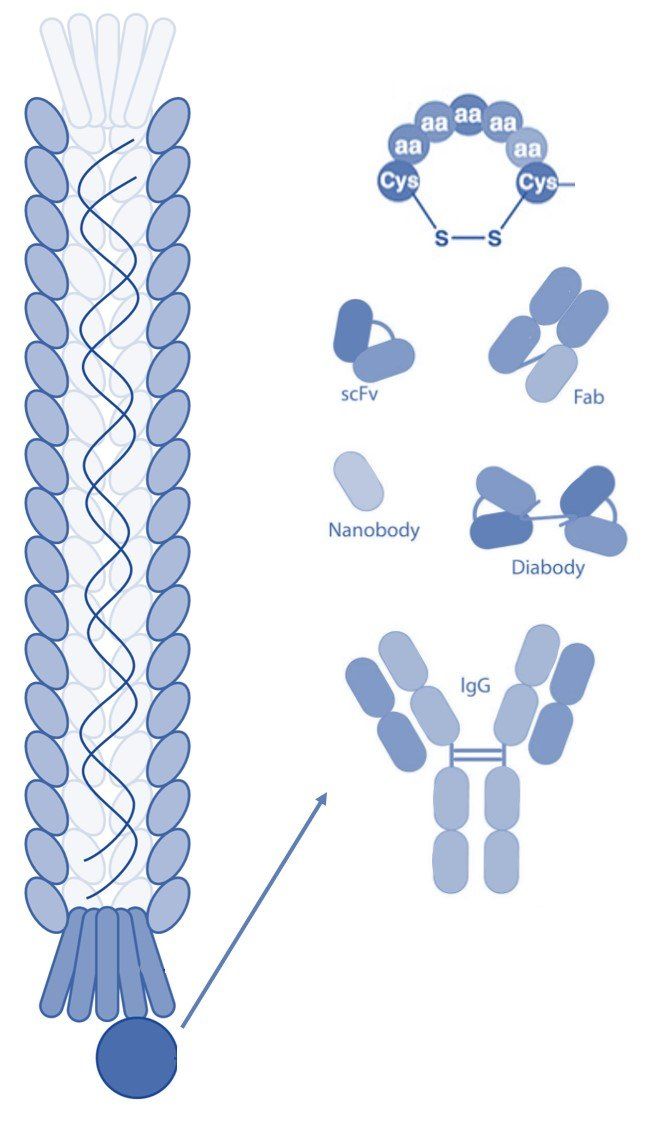
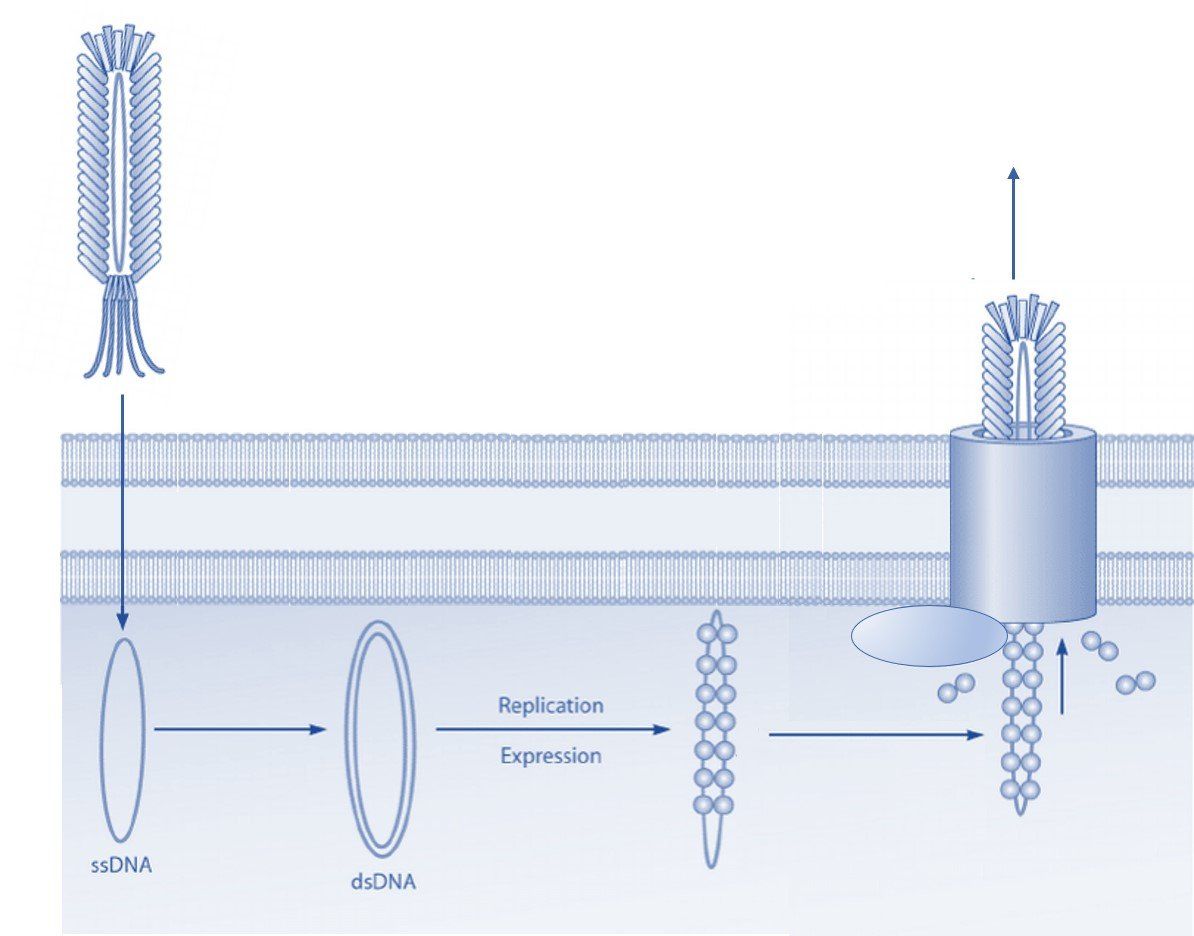
The Ff class of phages are structurally similar and 98% identical at the DNA level. These viruses are rod shaped and measure ~0.9 μm in length and ~65 Å in diameter. The Ff genome is approximately 6.4 kb (ssDNA) and encodes 11 proteins, of which five have structural roles. Two of these structural proteins are the minor coat protein III (pIII) and coat protein VIII (pVIII), which are the most commonly modified bacteriophage coat proteins in phage display technology [4]. Coat protein VIII is predominant on the phage surface and is usually found in more than 2300 copies distributed along the virion. Five copies of pIII are present at the distal end of the viral particle [5]. Both coat proteins are well exposed and are for this reason often used in phage display to present foreign peptides.
Coat protein III is utilized during infection of gram-negative bacteria such as E. coli. The F-pilus of the bacterium interacts directly with pIII, which induces retraction of the pilus and delivery of the phage genome into the host cell [5]. Although pIII is essential for infection of the bacterial cell, display on foreign peptides only minimally influences the ability of the phage to infect E. coli [6].
The phage biology of the Fd and M13 species is very similar. Both bacteriophages release viral progeny through the plasma membrane without lysis of the host cell [5]. As a result, propagation of filamentous bacteriophage in the lab is quick and straightforward and make them ideal for use in various phage display selection techniques.
T7 Phage
The T7 phage virion contains an icosahedral capsid with an inner diameter of 55 nm. A 29 nm tail is attached to the capsid, which is employed during infection of the host cell [7]. Thus, its morphology is vastly different from that of the filamentous bacteriophage. The T7 phages are able to infect most strains of E. coli. However, in contrast to the Ff class, the T7 phage follow a lytic lifecycle, which leads to destruction of the host cells [8]. The lytic lifecycle complicates propagation in the laboratory as it can be difficult to obtain sufficient numbers of virions. This makes the T7 phage less ideal as an expression method compared to the filamentous phage for use in phage display.
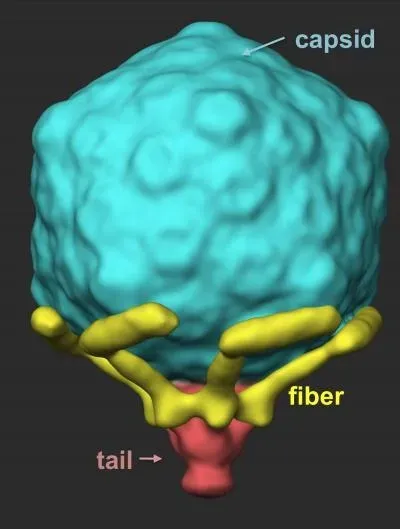
The T7 genome consists of approximately 40 kbp dsDNA that encodes 55 proteins. Most often, it is the gene 10 capsid protein that is used to display foreign peptides [7]. However, T7 phage display is restricted to smaller peptides as the genome cannot accommodate large inserts due to the limiting size of the viral capsid [9]. Filamentous phage do not suffer from this issue since a larger genome is easily accommodated during propagation by adding more coat proteins to the virion.
Summary
Filamentous and T7 phage may both be utilized in phage display technology. However, the Ff class of filamentous phage offer advantages in the laboratory. Specifically, these phage are easy to propagate and can accommodate a larger variety of displayed peptides.
References
[1] Smith, G. P. (1985). Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science. Vol 228(4705):1315-1317. doi: 10.1126/science.4001944
[2] Zacher, A. N., Stock, C. A>, Golden, J. W., Smith, G. P. (1980). A new filamentous phage cloning vector: fd-tet. Gene. Vol. 9(1-2):127-140. doi: 10.1016/0378-1119(80)90171-7
[3] Smith, G. P., and Petrenko, V. A. (1997). Phage Display. Chemical Reviews. vOL. 97(2):391-410. doi.org/10.1021/cr960065d
[4] Ping, X., Gosh. S., Gul A. R., Bhamore J. R., Park J. P., Park T. J. (2021). Screening of specific binding peptides using phage-display techniques and their biosensing applications. Trends in Analytical Chemistry. Vol. 137. doi.org/10.1016/j.trac.2021.116229
[5] Marvin, D A. (1998). Filamentous phage structure, infection and assembly. Current Opinions in Structural Biology. Vol. 8(2):150-158. doi.org/10.1016/S0959-440X(98)80032-8
[6] Derda R., Tang S.K., Whitesides, G. M. (2010). Uniform amplification of phage with different growth characteristics in individual compartments consisting of monodisperse droplets. Angewandte Chemie. Vol. 49(31):5301–5304. doi:10.1002/anie.201001143
[7] Molineux, I. J., Panja, D. (2013). Popping the cork: mechanisms of phage genome ejection. Nature Reviews Microbiology. Vol. 11(3):194–204. doi:10.1038/nrmicro2988
[8] Studier, F. W. (1969). The genetics and physiology of bacteriophage T7. Virology. Vol. 39(3):562–574. doi:10.1016/0042-6822(69)90104-4
[9] Danner, S., Belasco, J. G. (2001). T7 phage display: a novel genetic selection system for cloning RNA-binding proteins from cDNA libraries. Proceedings of the National Academy of Sciences of the United States of America U.S.A. Vol. 98(23):12954–12599. doi:10.1073/pnas.211439598
[10] Modified from ViralZone, SIB Swiss Institute of Bioinformatics. Permission to use ViralZone graphics in Wikimedia Commons kindly granted by SIB (legal(at)sib.swiss), 2021-02-01. - https://viralzone.expasy.org/resources/Inoviridae%5Fvirion.jpg on https://viralzone.expasy.org/113, CC BY-SA 4.0, https://commons.wikimedia.org/w/index.php?curid=99827306
[11] By Hu et. al. 2013. Science Express. Via https://www.eurekalert.org/pub_releases/2013-01/uota-vci011013.php - https://www.eurekalert.org/multimedia/pub/web/51639_web.jpg from https://www.eurekalert.org/multimedia/pub/51639.php, CC BY 4.0, https://commons.wikimedia.org/w/index.php?curid=99665366