Knowing When to Transition Your Pharmaceutical Discovery to Pre-IND Work: 6 Key Indicators
LeAnn Qi • March 19, 2024

Transitioning a pharmaceutical project from the discovery stage to pre-investigative new drug application (pre-IND) work is a momentous step, marking the shift from conceptual research to the prospect of new therapeutics. For pharmaceutical scientists, this is the stage that necessitates a meticulous evaluation process because each progression is laden with significant resource allocation and regulatory implications. Here are six critical checkpoints to confirm before taking the leap from drug discovery to the pre-IND phase.
1. Define a Clear Target Product Profile (TPP)
Before you advance to pre-IND work, a robust understanding of your Target Product Profile (TPP) is non-negotiable. The TPP delineates the characteristics of a therapeutic candidate required for it to be functional and competitive in the market. This profile dictates key information, ranging from therapeutic needs and clinical development to regulatory requirements and commercialization strategies.
Does your candidate satisfy these criteria? Would the data you've accumulated thus far support its regulatory approval and commercial success? Ensuring a seamless fit between your product profile and existing data sets the stage for informed decision-making and the efficient allocation of resources in the pre-IND phase.
2. Completion of Pilot Safety Studies
Safety evaluation is a linchpin in pharmaceutical development, and in the transition to the pre-IND phase, it's imperative that comprehensive pilot safety studies have been conducted. These preliminary studies serve as miniaturized versions of the broader toxicology assessments eventually required for FDA submission.
The insights from these studies guide the refined design of formal toxicology studies and help organizers understand the safety profile of their drug, a vital consideration in the decision to progress a molecule. It's at this point that potential red flags can emerge, necessitating further refinement or reconsideration of the developmental pathway.
3. Evident Signal of Efficacy
In the pre-IND phase, the requirement for financial investment tends to escalate significantly. Therefore, a clear and evident signal of efficacy becomes a critical element in favor of progressing further. This early demonstration of therapeutic potential, ideally in an in vivo model that closely mimics the clinical scenario intended for the drug, gives confidence to stakeholders—from investors to regulators—about the drug's viability.
A strong efficacy profile complements the established safety signal and becomes a key leverage in the compelling narrative required for successful partnerships and funding initiatives. Without it, the advancement may be significantly impeded.
4. Prepared Manufacturing Program and Scale-Up Considerations
The transition to pre-IND work necessitates that you have a solid manufacturing program in place. This program should be designed to produce the quantities and qualities of your candidate for upcoming clinical trials and eventual commercial-scale manufacturing. Along with this, an initial scale-up process should have been outlined, and modeling conducted to discern potential market prices.
The questions around manufacturing should be addressed thoughtfully. Where will it be done, and by whom? What are the potential bottlenecks, and how will they be mitigated? Understanding these aspects is vital for the successful execution of pre-IND work and subsequent phases of drug development.
5. A Competent and Cohesive Team
Pharmaceutical development is a complex and often multifaceted process. A well-organized and proficient team is indispensable for navigating the pre-IND phase effectively. This team should possess expertise that spans all dimensions of drug development, including regulatory affairs, toxicology, clinical development, formulation, and manufacturing.
Further, the presence of experienced consultants or advisory board members can offer invaluable guidance through the intricacies of the pre-IND and IND submission process. Having the right people in place ensures that the project benefits from comprehensive perspectives and seasoned judgment.
6. Engaged Key Opinion Leaders and Patient Insights
Informed collaboration with Key Opinion Leaders (KOLs) and the collection of patient insights are pivotal in the pre-IND phase, particularly as you move towards clinical trials. KOLs provide expert feedback on your drug's potential, clinical relevance, and prospective use. Likewise, direct patient input through surveys and focus groups can offer critical perceptions and preferences that shape your drug development trajectory.
Engaging with these stakeholders helps in aligning product development with identified needs, enhancing clinical trial design, and fine-tuning regulatory strategies. Ultimately, an understanding of how your product is perceived by those prescribing, taking, and paying for the medicine is imperative for effective positioning and adoption in the market.
Conclusion
By methodically addressing key components of drug development, you can approach this pivotal juncture with some confidence that you've measured the right things, thought about the right things and that your drug candidate is as ready as what makes sense for the arduous and rewarding path ahead. The pre-IND phase not only sets the tone for your drug's developmental pathway but acts as a blueprint for a product that could make a profound difference in patients' lives.
We at Cell Origins would love to collaborate with you in the discovery stage using phage display to develop drug candidates with superior drug profiles. Let's work together to bring novel therapeutics to market and improve patient outcomes.

The path of a compound from its identification in drug discovery to the commencement of formal human trials is marked by a crucial transitional phase - the pre-Investigational New Drug (pre-IND) work. This phase lays the groundwork for regulatory scrutiny and sets the trajectory for clinical success. For pharmaceutical researchers dedicated to the relentless pursuit of novel therapies, understanding the nuances of this transition is key to navigating the complex landscape of drug development.

As we celebrate Women's History Month at Cell Origins, we find it important to recognize the many women scientists who have contributed to the advancement of phage display technology. Phage display is an extremely powerful method for discovering peptides and antibodies for drug discovery, studying protein interactions, and elucidating structure-function relationships in a wide variety of biological contexts.

Phage display technology Phage display technology is a powerful tool used in drug discovery to develop peptides and monoclonal antibodies that bind specifically with high affinity to a target molecule. The technique involves genetically engineering bacteriophages to display foreign peptides or antibodies on a viral coat protein to create a filamentous fusion phage. Phage display libraries are collections of such phages that typically contain 10^9-10^10 different phage clones, making them a powerful tool for screening and discovering new targeting molecules. Due to their flexible design and large-scale production capabilities, phage display libraries have become an important tool in the drug discovery process, allowing scientists to rapidly screen for potential new drug targets and candidate compounds. Additionally, phage display technology has been successfully employed in several other areas of scientific research, including immunology and protein engineering. Here we give a brief overview of the key advantages of phage display in drug discovery. Phage Display is Highly Versatile Phage display can be used for a variety of applications at different stages in drug development including peptide and antibody discovery, drug candidate screening, and lead optimization. Additionally, phage display libraries can be screened using a variety of different biopanning methodologies depending on the needs of the researcher. Biopanning involves iterative rounds of affinity selection against a target molecule, cell, or tissue to select phages that display peptides or antibodies with certain desired binding and pharmacokinetic characteristics. During biopanning, a phage library is exposed to the target molecule, and unbound or weakly bound phages are removed by washing, after which bound phage particles are collected by elution. Phage display biopanning is versatile because almost any type of peptide or antibody library can be screened against any type of target. For in vitro biopanning experiments, the phage display library is typically affinity selected against recombinant proteins or other isolated molecules. This type of selection often results in the identification of high-affinity binders. However, many studies have shown that peptides and antibodies selected by in vitro phage display often fail to exhibit optimal binding in live tissues and whole organisms and demonstrate less-than-ideal pharmacokinetic properties. Instead, more researchers are recognizing that phage display selections against cells and tissues (in situ) and in live animals or patients (in vivo) offer advantages regarding the identification of ligands that show high binding affinity, minimal off-target binding, and optimal pharmacokinetics. This is especially useful when interrogating a complex biological process that cannot easily be replicated in vitro, such as differentially expressed target proteins of varying lengths and accessibility, as well as ones that have different binding partners depending on the tissue, as is often observed in normal vs. tumor tissues. At Cell Origins , we are pioneers in utilizing multi-tier phage display techniques to develop peptides and monoclonal antibodies that show desirable binding kinetics and pharmacokinetics. Our platform ensures that lead compounds maintain their high binding affinity and specificity while having optimal pharmacokinetics and minimal off-target binding. Phage Display is High Throughput The phage display platform is highly efficient and can screen billions of different peptides or antibodies in a single day. This high throughput capacity makes phage display an attractive technology in drug discovery. Practically, this means that phage display selections can be used to screen large libraries in a matter of a few days and that attractive drug candidates can be identified in as little as a few weeks after next-generation sequencing and bioinformatic analysis have been completed. Furthermore, using filamentous phage to display desirable ligands offer the ability to screen and analyze candidates in a high throughput and cost-effective manner that is not possible with individual soluble peptides and antibodies. As such, recombinant DNA technology, rapid propagation in E. coli, and the development of easy tagging methods allow kinetic analyses to be carried out post-selection in a much more rapid manner. While the synthesis of peptide sequences and the production of monoclonal antibodies typically take weeks to complete, the production of phage for kinetic analyses can be carried out in a few days. Phage Display is Cost-Effective Phage display screening is among the least expensive investigation techniques available for drug discovery. Compared with other technologies, phage display selections typically require fewer resources, reagents, and consumables while generating targeting ligands with high-affinity and desirable binding properties in a short amount of time. Additionally, phage libraries are known to be highly robust and can be stored for extended periods of time without a significant loss in infectivity and usability. The cost-effectiveness and versatility of phage display technology allow the researcher to simultaneously interrogate several types of antibody and peptide libraries against potential biomarkers under different conditions. Such a multiplexing approach lowers the overall cost of drug discovery and further provides an opportunity to add additional levels of competition and complexity to the phage display selection to more rapidly generate ligands with optimal properties. Summary Phage display provides a versatile and powerful tool to rapidly identify high-affinity ligands in a cost-effective manner. At Cell Origins , we are experts in phage display with extensive experience in generating peptides and antibodies that bind difficult and complex targets both in vitro and in vivo. Our multi-tier phage display selection platform is proven to result in ligands that exhibit high affinity, and excellent pharmacokinetics while showing minimal off-target binding. Please contact us to learn more about how our unique phage display platform can accelerate your drug discovery. Learn More Biopanning https://en.m.wikipedia.org/wiki/Biopanning Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. https://www.frontiersin.org/articles/10.3389/fimmu.2020.01986/full Phage-displayed peptides targeting specific tissues and organs. https://www.tandfonline.com/doi/full/10.1080/1061186X.2018.1531419 Phage display--a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3656071/ Phage display antibody libraries: A robust approach for generation of recombinant human monoclonal antibodies. https://www.sciencedirect.com/science/article/abs/pii/S0141813019330855?via%3Dihub Basics of Antibody Phage Display Technology. https://www.mdpi.com/2072-6651/10/6/236 Phage display screening of therapeutic peptide for cancer targeting and therapy. https://link.springer.com/article/10.1007/s13238-019-0639-7 Phage Display Libraries: From Binders to Targeted Drug Delivery and Human Therapeutics. https://link.springer.com/article/10.1007/s12033-019-00156-8 Advancement and applications of peptide phage display technology in biomedical science. https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-016-0223-x

Movember is a month-long campaign that raises awareness about men's health issues, such as prostate cancer , testicular cancer , and mental health. At Cell Origins , we participate in Movember by highlighting the importance of prostate cancer research and how the correct use of cell lines play a crucial role. Prostate Cancer Prostate cancer is one of the most common types of malignancies, affecting an estimated 1 in 7 men during their lifetime. In many cases, prostate cancer grows slowly and remains confined to the prostate gland. However, some types of prostate cancer are aggressive and can metastasize quickly to other parts of the body. Early diagnosis and treatment are key for improving survival rates among men with prostate cancer. Prostate cancer is typically diagnosed by performing a digital rectal exam and prostate-specific antigen ( PSA ) test. Often additional imaging, such as transrectal ultrasound and magnetic resonance imaging (MRI), as well as biopsy procedures are needed to confirm a diagnosis. Several treatment options for prostate cancer are available, and these most often include surgery, chemotherapy, radiation, immune, and hormone therapy. Initially, a prostatectomy , which involves partial or complete removal of the prostate, is performed in combination with other treatments, most often radiation therapy and androgen deprivation therapy ( ADT ). Chemotherapy using docetaxel or the hormone drug abiraterone is typically added to the treatment regimen when prostate cancer is diagnosed at later stages. Cell Lines are Important Tools in Prostate Cancer Research Cell lines are essential tools in prostate cancer research, allowing scientists to study and explore the cellular mechanisms of the disease. Such studies have led researchers to discover new chemo- and immuno-therapies, develop more effective diagnostic methods, and elucidate central genomic, transcriptomic, and proteomic signatures. However, continuous research is needed to further the development of more efficient diagnostic and therapeutic methods. Unfortunately, cell line contamination, misidentification, and phenotypic drifting due to changes in cell culturing can greatly impact the reproducibility and accuracy of research data. The Use of Contaminated and Misidentified Cell Lines The persistent existence of contaminated and misidentified cell lines (CMCL) within the scientific community is a major problem that has significant implications for scientific research [ 1 , 2 , 3 , 4 ]. Such CMCLs have unknowingly been utilized in scientific studies for decades, which has likely significantly impacted the advancement of cancer research [ 5 ]. For example, HeLa contamination was first noted in the 1960s and continues to be an issue today [ 6 ]. Cell line contamination is usually caused by human error, such as using improper sterile techniques when handling the cells. Misidentification can happen for a number of reasons, but it is often due to cross-contamination between different cell lines. Both contamination and misidentification can introduce significant errors and lead to inaccurate conclusions. Additional problems of over-cultivating cell lines are also beginning to be addressed within the literature [ 7 , 8 ]. Culturing cell lines for too long with various unintentional selective pressures such as temperature, pH changes, and nutrient availability results in phenotypic drifting. A myriad of altered gene expression profiles that leads to diverse morphologies and biomarker expression can then impact the reproducibility of your experiments, as well as the accuracy of your data. Thus, replicating experiments becomes difficult if not impossible depending on how many variations in cell culture have taken place. Currently, it is estimated that CMCLs and phenotypically drifted cell lines are used in 20-30% of published data [ 9 ]. Given these concerns, it is critical that researchers are aware of the risks associated with using CMCLs and take steps to prevent their continued use. Probing Three Tiers of Biology Unfortunately, there is currently no widely available and standardized method that combines cell line verification and monitoring of phenotypic changes. Short tandem repeat (STR) analysis is the most common method of cell line identification and was standardized by the American National Standards Institute ( ANSI ) and American Type Culture Collection ( ATCC ). However, this method fails to detect mutations outside of the amplicon regions as well as changes in cellular phenotypes. The growing need to develop new methods for the identification and phenotypic characterization of cell lines was identified by the National Institute of Health ( NIH ). Cell Origins was awarded a small business innovation research ( SBIR ) grant from the NIH in 2019 to address this need. Specifically, we are developing a phage-based method to probe changes to the three tiers of biology (genomic, transcriptomic, and phenotypic) that occur as a result of altered cell culture conditions such as prolonged passaging. Phage Clones Detect Changes in Culture Conditions of a Prostate Cancer Cell Line At Cell Origins, we have used phage display technology to develop phage clones that can discern the phenotypic changes resulting from prolonged culturing of the prostate cancer cell line, LNCaP . Phage display is a technology used to identify peptides and antibodies that bind to the desired target molecule. For our product, we used phage display technology to identify phage clones that specifically bind to LNCaP biomarkers that are differentially expressed at different passage numbers of cultures. Combined with genomic and transcriptomic analysis, this allows us to discern if the cells have significantly drifted and can no longer be considered the same as the originally purchased cells. Currently, our technology is limited to the prostate cancer cell line LNCaP. However, we are busy in our research and development laboratory developing phage clones that can identify and detect changes to the most common cell lines distributed by the ATCC. We envision that our phage-based technology will be used in research laboratories to prevent the use of CMCLs in experiments and to ascertain phenotypic characteristics. We suggest that researchers periodically evaluate their cell lines in this manner. If the researcher is not amenable to the removal of questionable or over-cultured cell lines from their laboratory, they may instead report the genomic, transcriptomic, and phenotypic characteristics along with the experimental results. This added information will aid in the scientific community’s ability to evaluate published data. We Are Looking for Beta-Testers The Cell Origins team is excited to announce the development of our technology to authenticate cell lines and determine phenotypic drift. This technology will enable the standardization of mammalian cell culture and will help researchers ensure that their data is reproducible. We are currently looking for beta testers for our product, and we hope that you will consider joining us. Our technology is able to identify and monitor cultured cell lines for phenotypic drift that results from changes in culturing so that you can ensure the quality of your data. To become a beta tester, please contact us . We look forward to working with you! Summary At Cell Origins, we celebrate Movember by raising awareness about the importance of continued prostate cancer research and how cell lines play a key role in this. Unfortunately, cell line contamination, misidentification, and phenotypic drifting due to human errors and changes in cell culturing can impact the reproducibility and accuracy of data. Cell Origins is working to improve the accuracy of data in prostate cancer research by developing a new technology that can identify cell lines and ascertain phenotypic drifting. This is important work that has the potential to advance prostate cancer research to improve the outcomes for patients suffering from this disease. Please contact us if you are interested in beta testing our technology and learning more. Further Readings Movember https://us.movember.com/ American Cancer Society https://www.cancer.org/cancer/prostate-cancer.html National Cancer Institute https://www.cancer.gov/types/prostate Prostate Cancer Foundation https://www.pcf.org/ Center for Disease Control and Prevention https://www.cdc.gov/cancer/prostate/basic_info/index.htm References [1] Horbach SPJM, Halffman W. The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PLoS One. 2017 Oct 12;12(10):e0186281. doi: 10.1371/journal.pone.0186281. PMID: 29023500; PMCID: PMC5638414. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0186281 [2] Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010 Jul 1;127(1):1-8. doi: 10.1002/ijc.25242. PMID: 20143388. https://onlinelibrary.wiley.com/doi/full/10.1002/ijc.25242 [3] Freedman LP. Know Thy Cells: Improving Biomedical Research Reproducibility. Sci Transl Med. 2015 Jul 1;7(294):294ed7. doi: 10.1126/scitranslmed.aac7112. PMID: 26136474. https://www.science.org/doi/10.1126/scitranslmed.aac7112 [4] Vaughan L, Glänzel W, Korch C, Capes-Davis A. Widespread Use of Misidentified Cell Line KB (HeLa): Incorrect Attribution and Its Impact Revealed through Mining the Scientific Literature. Cancer Res. 2017 Jun 1;77(11):2784-2788. doi: 10.1158/0008-5472.CAN-16-2258. Epub 2017 Apr 28. PMID: 28455420. https://aacrjournals.org/cancerres/article/77/11/2784/616254/Widespread-Use-of-Misidentified-Cell-Line-KB-HeLa [5] Freedman LP, Cockburn IM, Simcoe TS. The Economics of Reproducibility in Preclinical Research. PLoS Biol. 2015 Jun 9;13(6):e1002165. doi: 10.1371/journal.pbio.1002165. Erratum in: PLoS Biol. 2018 Apr 10;16(4):e1002626. PMID: 26057340; PMCID: PMC4461318. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002165 [6] Lavappa KS, Macy ML, Shannon JE. Examination of ATCC stocks for HeLa marker chromosomes in human cell lines. Nature. 1976 Jan 22;259(5540):211-3. doi: 10.1038/259211a0. PMID: 1250349. https://www.nature.com/articles/259211a0 [7] Hughes P, Marshall D, Reid Y, Parkes H, Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques. 2007 Nov;43(5):575, 577-8, 581-2 passim. doi: 10.2144/000112598. Erratum in: Biotechniques. 2008 Jan;44(1):47. PMID: 18072586. https://www.future-science.com/doi/full/10.2144/000112598?rfr_dat=cr_pub++0pubmed&url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org [8] Masters JR, Stacey GN. Changing medium and passaging cell lines. Nat Protoc. 2007;2(9):2276-84. doi: 10.1038/nprot.2007.319. PMID: 17853884. https://www.nature.com/articles/nprot.2007.319 [9] Lacroix M. Persistent use of "false" cell lines. Int J Cancer. 2008 Jan 1;122(1):1-4. doi: 10.1002/ijc.23233. PMID: 17960586. https://onlinelibrary.wiley.com/doi/full/10.1002/ijc.23233
Cell Origins is a woman-owned business founded by Jessica Newton-Northup, Mette Soendergaard, and Leann Kuhlmann Qi in 2018. Years earlier, Mette Soendergaard and Jessica Newton-Northup formed and maintained a collaborative relationship while at the University of Missouri , the birthplace of phage display technology. Here, they developed a passion for phage display and the development of phage-based methods that they now bring to Cell Origins. Our Origin and Evolution At Cell Origins, our passion for phage-based solutions drives us to develop innovative products and services that address the growing challenges facing the biomedical research community. We started as a company to meet a growing need identified by the National Institute of Health ( NIH ) to develop new methods for the identification and phenotypic characterization of eukaryotic cells. Cell Origins was awarded a small business innovation research ( SBIR ) grant from the NIH in 2019 to develop a phage-based method to discern the genomic, transcriptomic, and phenotypic changes to the cell biology of human cells. Thus, we named our company Cell Origins. The Origins of Cells Cell lines are an essential tool for scientific research, allowing scientists to study and explore a wide range of diseases and cellular mechanisms. However, cell line contamination, misidentification, and phenotypic drifting due to changes in cell culturing can greatly impact the reproducibility and accuracy of data. In fact, recent studies have found that up to 20-30% of all eukaryotic cell lines in use may be contaminated or misidentified , leading to significant variability in data. To address this issue, we are developing a new phage-based laboratory technology for improving data reproducibility in cell line experiments by rapidly and accurately identifying cell lines and characterizing cell phenotypes. Specifically, the method is able to identify the origin of cells and monitor living cells for phenotypic drift that results from contamination and changes in culture. The technology combines the characterization of cell-type biomarkers with the quantification of RNA transcripts and short-tandem repeat (STR) sequencing. Our Phage Display Origins Phage display technology was first developed by Dr. George Smith at the University of Missouri in 1985. He was later awarded the Nobel Prize in chemistry for his work on "the phage display of peptides and antibodies". Phage display technology involves using filamentous bacteriophages to display peptides or antibodies on their surface, allowing for the selection and screening of specific ligand molecules with desired characteristics. In phage display selection experiments, also known as biopanning , a peptide or antibody library is incubated with a target molecule, living cells, or tissue under conditions that allow only specific interactions between the target and the displayed peptide or antibody. After washing away unbound phage clones, bound phages are collected by elution and amplified in E. coli . Peptide or antibody sequences that bind to the target with high affinity are identified by DNA sequencing and bioinformatic analysis to generate a short list of interesting leads. In the years since its development, phage display has been used to discover novel peptides and antibodies for a variety of biomedical applications. Atezolizumab and Necitumumab are antibodies that target PD-L1 and the epidermal growth factor receptor (EGFR), respectively, and have been approved by the Food and Drug Administration ( FDA ) to treat various cancers. Recently, the short peptide Pegcetacoplan was approved by the FDA for the treatment of Paroxysmal Nocturnal Hemoglobinuria (PNH). The founders of Cell Origins were trained at the origin of phage display, at the University of Missouri , and it was there that their passion for using phage display to develop cancer-targeting peptides and antibodies began. While at the University of Missouri, the scientists at Cell Origins pioneered several phage display selection and analysis methods. Their passion is now used at Cell Origins to help researchers in drug discovery achieve their scientific goals. Cell Origins offers a variety of phage display services, including phage display selections , screening, premade and custom peptide and antibody libraries, and consulting on research strategies to deliver peptides and antibodies with superior binding properties and pharmacokinetics. At Cell Origins We Are Unique Cell Origins is a unique company in the phage display space in that we offer a custom approach that combines multi-tier selections. We work closely with our clients to gain an in-depth understanding of the molecular target and disease mechanisms. From that, we develop a detailed and custom research strategy that ensures the selection of peptide or antibody leads with specific and high-affinity binding as well as the desired pharmacokinetic properties. We specialize in difficult targets that others often have failed such as small epitopes, and biomarkers with minimal expression differences in normal versus disease-associated tissues. Further, our skilled scientists are experts in ensuring that your peptide or antibody maintains its affinity and specificity in vivo to deliver excellent targeting and pharmacokinetics. Contact us to learn more about our unique approaches to phage display.

October is Women’s Business Month in many states, and we at Cell Origins would like to take this opportunity to celebrate the women-owned businesses and women entrepreneurs that play a pivotal role in contributing to the economy and strengthening our local communities. Here at Cell Origins, we are proud to be a women-owned business that was founded by Mette Soendergaard (Ph.D.), Jessica Newton Northup (M.S.), and LeAnn Kuhlmann Qi (M.B.A) in 2018. Cell Origins provides researchers with critical phage display laboratory services including peptide and antibody selections , screening, premade and custom phage display libraries, as well as consulting on research strategies related to peptide and antibody discovery. Our mission is to provide the highest quality services by employing non-traditional phage display strategies such as multi-tier selections and proprietary high-throughput analysis tools so that our clients can achieve their research goals in drug discovery. We are committed to providing high-quality peptides and antibodies that exhibit outstanding binding characteristics and optimal pharmacokinetics. The Importance of Women-Owned Businesses Women-owned businesses are one of the fastest-growing segments of the economy, and they play a vital role in creating jobs, driving innovation, and growing our local economies. According to the National Association of Women Business Owners ( NAWBO ), there are more than 11 million women-owned businesses in the United States that generate nearly $1.8 trillion in revenue and employ nearly 9 million people. Successful Women Entrepreneurs in Science Successful women entrepreneurs in science are becoming an increasingly important part of the business landscape. With the rise of female entrepreneurs, there is a growing need for high-quality mentor networks that can support other female entrepreneurs as they navigate unique challenges and obstacles, such as work-life balance and securing adequate funding. Despite some persisting barriers, women in STEM continue to be a driving force in this rapidly changing landscape.
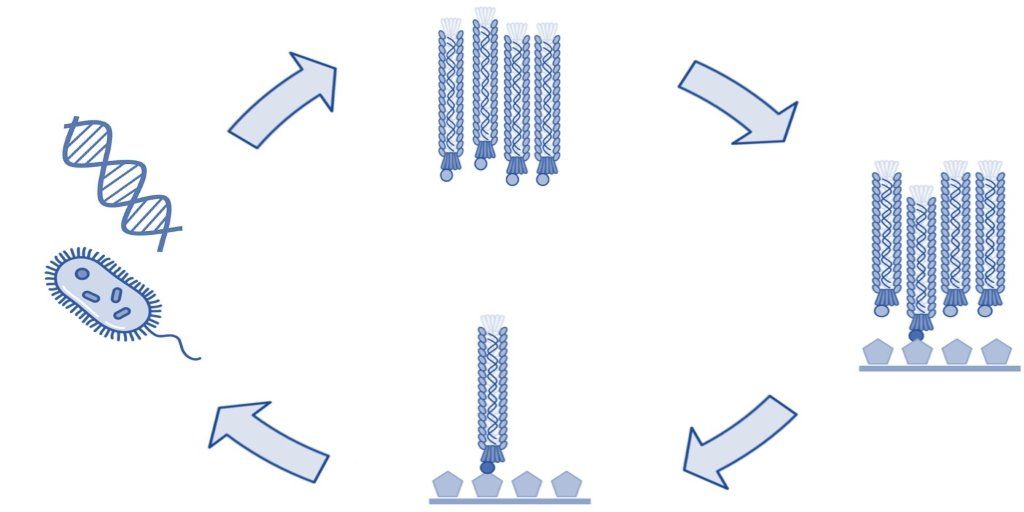
Phage display technology is a laboratory technique that employs genetically modified filamentous bacteriophages to display foreign peptides, antibodies, or other proteins. The phage particles are assembled into phage display libraries, from which researchers can select and identify peptide sequences or antibodies that bind with high affinity and specificity to a target molecule. Phage display technology has been used for many purposes, especially in drug discovery for the development of therapeutic antibodies and peptides . Biopanning Biopanning of phage libraries is the most common method of selecting and identifying peptides and antibodies that bind to a target antigen. A traditional biopanning procedure includes the incubation of a phage library with the target of interest followed by the collection of bound phages by elution. Each biopanning protocol contains the following key steps, which are described in more detail below. Incubation of the phage display library with the target of interest (affinity selection). Removal of unbound phage by washing. Collection of bound phage by elution. Amplification of collected phage. Repetition of steps 1-4 for typically a total of four rounds. Identification of collected phage by DNA sequencing. While phage display biopanning is a relatively simple and efficient technique, it requires careful optimization of several parameters in order to achieve the best results. These include the choice of the type of phage library, phage and target molecule concentration, cell type (tumor cells, bacterial cells, etc.), incubation conditions such as temperature and time, as well as the amplification method of collected phage. Affinity Selection The first step of a biopanning process is often referred to as affinity selection since this step allows phage-displayed peptides or antibodies with affinity for the target to bind. Often, the target molecule is an immobilized recombinant protein. However, carbohydrates, lipids, and other biomolecules can also be used. Additionally, whole cells, tissues, and organisms (experimental animals, human patients, etc.) have been used with great success in biopanning protocols to identify binding peptides and antibodies with high affinity and specificity. For affinity selection, it is important to consider the stringency of the selection, which can be modified by changing the concentration of either the phage libraries or the target antigens. For example, to select high-affinity ligands binding competition between the different phage clones may be increased by raising the concentration of the phage library or by lowering the concentration of the target molecule. Such stringent conditions can significantly increase the likelihood of identifying desirable phage clones. Removal of Unbound Phages Unbound phages are typically removed by extensive washing using a buffer with a low concentration of a detergent. Weakly bound phages, which are typically undesirable in the phage display selection, can be removed by increasing the concentration of detergent or adding additional washing steps. Elution of Bound Phages In standard biopanning procedures, bound phages are collected by elution using detergents, changes in pH, or other methods of disrupting the non-covalent interactions between the phage and target moiety. Using such elution methods generally results in the collection of the vast majority of bound phages. However, phage clones with high binding affinity may not be sufficiently eluted by disruption of the non-covalent interactions. Thus, trypsin digestion has become prevalent and ensures equivalent elution of the bound phages. Amplification of Phages After each selection round, eluted phages are usually amplified and used in subsequent selections. Amplification of collected phages can be done in different ways and depends on the type of vector used for the phage library. Bacteriophage vectors such as the fUSE5 library are most often amplified by the transduction of E. coli by the phage particle. Transformation by E. coli electroporation of the bacteriophage vector is typically complicated by the large size of the filamentous phage genome. Thus, this method often results in low transformation rates and valuable clones may be lost in this manner. Phagemid vectors are significantly smaller than bacteriophage vectors and are, therefore, more easily transformed into E. coli cells. However, for the propagation of phage particles, a helper phage is required to ensure the assembly and release of the viral progeny. Amplification of selected phage in E. coli is an easy and efficient method. However, the infection of E. coli can be influenced by the phage-displayed peptide or antibody. The infection of E. coli by filamentous bacteriophage is mediated by the interaction between the phage coat protein III (pIII) and the bacterial F pili. The use of the popular pIII-display libraries can thus lead to a change in the infection efficiency and create a selection bias towards certain phage clones. This selection bias may be overcome by using PCR to amplify the foreign oligonucleotide sequences encoding the displayed peptide or antibody. The PCR amplicons can then be inserted back into the bacteriophage vector or phagemid for subsequent transformation and propagation in E. coli . Repeating the Selection Rounds Most biopanning procedures include multiple rounds of affinity selection. Typically, four rounds of biopanning against the target antigen are employed to ensure sufficient selection pressure. However, fewer rounds of affinity selection may be used if the protocol includes deep next-generation sequencing of the eluted phages and bioinformatic analysis to sort the identified sequences. Different types of affinity selections may also be used in biopanning to identify phage clones with specific binding characteristics. For example, phage display selection against a recombinant protein may be combined with a selection round using a cell line that expresses the target protein on the cell surface. Such multi-tier biopanning strategies increase the likelihood of selecting peptides and antibodies that have optimal binding properties both in vitro and in vivo . Identification of Phage Clones Eluted phages from affinity selections are identified by DNA sequencing of the foreign inserts that encode the phage-displayed peptides or antibodies. Typically, only the phage clones from the last round of affinity selection are subjected to DNA sequencing. However, it can be an advantage to sequence the phage mixtures after each round of selection. Doing so allows the researcher to follow the efficiency of the phage display procedure since phage particles with binding affinity for the target should become more populous as the selection proceeds. In addition, it is advantageous to identify unbound phages that are removed from the selection by washing. This step helps identify target-unrelated peptides (TUP) and antibodies that may otherwise be incorrectly identified as potential leads. It may also identify so-called over-growers that exhibit a propagation advantage by infecting E. coli cells at a comparatively higher rate. Summary Biopanning is the most common phage display strategy used to select antibodies and peptides with high affinity and specificity for a target molecule. Biopanning is done in several steps that include affinity selection, removal of unbound phages by washing, elution and amplification of bound phages, as well as DNA sequencing to identify selected phage clones. While biopanning can be straightforward, several considerations are important, such as the choice of peptide library or antibody library, the affinity selection conditions, and the extent of DNA sequencing. When taking these considerations into account, biopanning can be a powerful tool for discovering therapeutic antibodies and peptides. Do you need a custom peptide, antibody, or biopanning strategy for your research? Cell Origins is the perfect partner for advanced biopanning strategies. Our scientists have years of experience in phage display technology and will work with you to find the best solution for your needs. With our multi-tier phage display selection process, we can provide you with the most optimal peptides and antibodies for your research with superior binding characteristics and pharmacokinetics. Contact us today to discuss your research needs. Learn More Ehrlich GK, Berthold W, Bailon P. Phage display technology. Affinity selection by biopanning. Methods Mol Biol. 2000;147:195-208. doi: 10.1385/1-59259-041-1:195. PMID: 10857097. https://pubmed.ncbi.nlm.nih.gov/10857097/ Li Y, Liu M, Xie S. Harnessing Phage Display for the Discovery of Peptide-Based Drugs and Monoclonal Antibodies. Curr Med Chem. 2021;28(40):8267-8274. doi: 10.2174/0929867327666201111144353. PMID: 33176631. https://www.eurekaselect.com/article/111400 Lim CC, Woo PCY, Lim TS. Development of a Phage Display Panning Strategy Utilizing Crude Antigens: Isolation of MERS-CoV Nucleoprotein human antibodies. Sci Rep. 2019 Apr 15;9(1):6088. doi: 10.1038/s41598-019-42628-6. PMID: 30988390; PMCID: PMC6465254. https://www.nature.com/articles/s41598-019-42628-6 McGuire MJ, Li S, Brown KC. Biopanning of phage displayed peptide libraries for the isolation of cell-specific ligands. Methods Mol Biol. 2009;504:291-321. doi: 10.1007/978-1-60327-569-9_18. PMID: 19159104; PMCID: PMC4053471. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4053471/pdf/nihms326434.pdf Perera DJ, Scully C. The phage display of peptides and proteins: a practical approach. Curr Protoc Mol Biol. 2014;105:2.44.1-2.44.12. doi: 10.1002/0471142727.mb0244s105. PMID: 2448 https://en.m.wikipedia.org/wiki/Biopanning
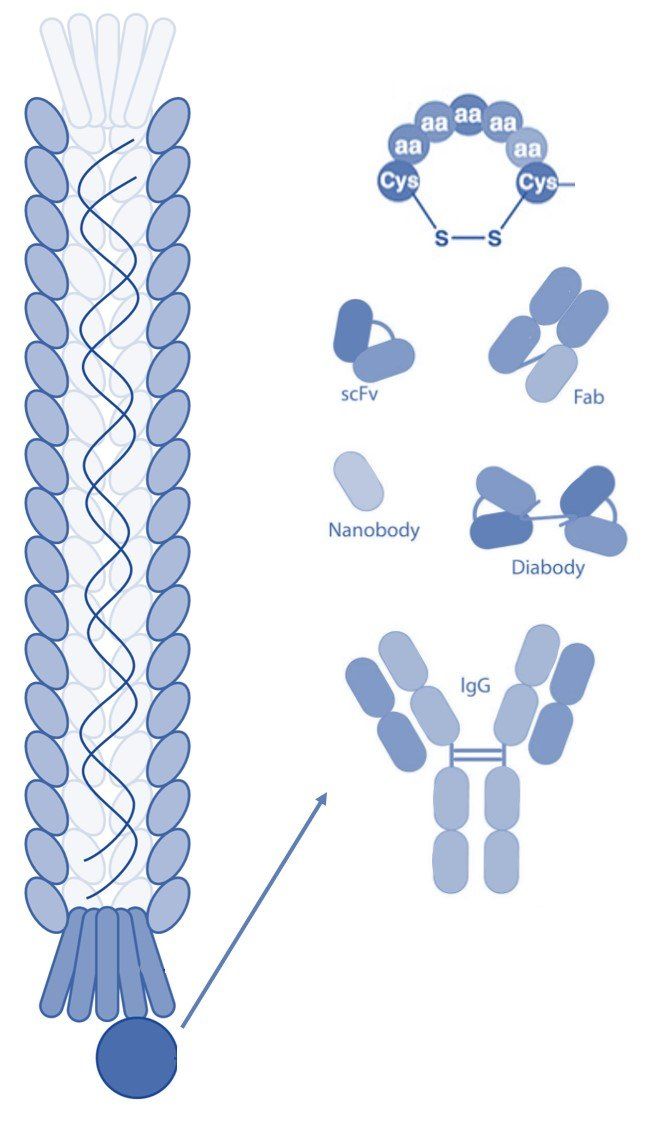
Phage display technology is used to select and identify peptides, monoclonal antibodies, or proteins that bind to a specific target molecule. The technique is commonly used in the pharmaceutical industry to develop new drugs such as peptide ligands and antibodies, and in immunology research to study antibody-antigen interactions. The process of phage display technology involves creating a filamentous fusion phage that displays a random foreign peptide or antibody linked to a phage coat protein. The filamentous phage are then assembled into a phage display library of random peptide sequences or antibodies. These phage display libraries are typically used to select peptide ligands, monoclonal antibodies, or antibody fragments with specific characteristics and binding properties, or used in protein engineering. Here we give an overview of the most common uses of peptide and antibody phage display libraries. Phage Display Peptide Libraries Peptide phage display technology is a powerful technique that can be used for a wide variety of applications in research and drug discovery. Peptides can be used directly as drugs to elicit therapeutic effects or to carry cargo to target biomarkers of disease. The small size of peptides offers several advantages as these molecules can extravasate into tissues and can be readily cleared from the body. This is a particular advantage for peptides used in the radiotherapy of cancer where the payload must be delivered directly to the target cells and be rapidly cleared to avoid off-target tissue damage. Linear Peptides Both linear and cyclic peptide sequences can be selected using phage display technology and each has its distinct advantages and disadvantages. Linear phage display peptide libraries have been available for decades and are typically cheaper compared to their cyclic counterparts. Additionally, the synthesis of soluble linear peptides is most often easier and more cost-effective, making them a more convenient option for researchers. However, they can have limited bioavailability due to their rapid degradation in the body, which can limit their efficacy as a drug unless they are substantially modified. Cyclic Peptides Several novel phage display libraries of cyclic peptides have been developed over the last decade. Such phage-displayed peptide libraries are typically created by inducing disulfide bond formation between cysteine residues, or by using a scaffolding system. Cyclic peptides offer better stability due to their non-natural structure. They also typically offer higher target binding affinity and specificity as a result of their rigid structure. This makes them ideal for applications where high potency or selectivity is required, such as cancer treatments or neurological disorders like Alzheimer's disease. However, the complex synthesis process involved in creating cyclic peptides can be costly and time-consuming, which may make them less desirable for some researchers. Phage Display Antibody Libraries Antibody phage display libraries are used in drug discovery and in many research applications. These libraries are often used to screen for antibodies with high binding affinities and specificities for a wide range of targets, including proteins, peptides, small molecules, and whole cells. Full-Length Monoclonal Antibodies Full-length monoclonal antibodies have become one of the most important classes in biopharmaceuticals. In fact, therapeutic antibodies continue to dominate markets with six out of ten top-selling drugs belonging to this class. The development of antibody phage display libraries as an alternative technique to the traditional hybridoma technology has accelerated the discovery of monoclonal antibodies. Fully human antibodies derived from large Ig gene repertoires can be assembled in phage display libraries and used to select full-length antibodies with desirable characteristics. The use of a phage display antibody library offers advantages over murine-derived monoclonal antibodies produced by the traditional hybridoma technology that often exhibit limited therapeutic efficiency due to accelerated clearance and allergic reactions. Antibody Fragments The development of smaller recombinant antibody fragments, such as variable domain (Fv), single-chain variable domain (scFv), diabodies (bivalent scFvs), fragment antigen binding (Fab), and heavy-domain camelid antibody fragments (nanobodies) have helped further advance antibody phage display. These antibody fragments are more amenable to expression than full-length antibodies that require the assembly of four polypeptide chains and extensive disulfide bond formation. Phage display libraries of human rearranged V-gene repertoires can be constructed from naïve or immunized donors. An advantage of the latter is that they are made from patients who have been immunized with an antigen of interest or who carry a disease, such as a specific cancer type. Thus, a phage display library of this type typically contains a biased antibody repertoire toward a specific target molecule, which can lead to high-affinity antibodies compared to monoclonal antibodies derived from naïve phage display libraries. On the contrary, a naïve phage display library that is combined from a set of healthy donors contain a more diverse repertoire that can be used to select antibody fragments against an unlimited range of target molecules. Antibody libraries can also be designed synthetically by randomization of the complementarity-determining regions (CDR). For this, random oligonucleotides are synthesized and inserted into the CDRs without disrupting the folding of the V regions thus generating a diverse and completely "naïve" antibody library. Summary Phage display technology is a powerful tool that can be used to select peptides and antibodies for use in drug discovery. Peptides and antibodies are vastly different molecules and each offers its own advantages and disadvantages, especially regarding tissue extravasation, immunogenicity, specificity, and binding affinity. Cell Origins offers premade and custom linear and cyclic phage display peptide libraries as well as antibody phage display libraries with proven high diversity that have resulted in the selection of ligands with optimal binding characteristics. Please contact us to learn more about how we can help with your phage display selections. Learn More Alfaleh MA, Alsaab HO, Mahmoud AB, Alkayyal AA, Jones ML, Mahler SM, Hashem AM. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front Immunol. 2020 Aug 28;11:1986. doi: 10.3389/fimmu.2020.01986. PMID: 32983137; PMCID: PMC7485114. https://www.frontiersin.org/articles/10.3389/fimmu.2020.01986/full Andrieu J, Re F, Russo L, Nicotra F. Phage-displayed peptides targeting specific tissues and organs. J Drug Target. 2019 Jun-Jul;27(5-6):555-565. doi: 10.1080/1061186X.2018.1531419. Epub 2018 Oct 24. PMID: 30281393. https://www.tandfonline.com/doi/full/10.1080/1061186X.2018.1531419 Bazan J, Całkosiński I, Gamian A. Phage display--a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Hum Vaccin Immunother. 2012 Dec 1;8(12):1817-28. doi: 10.4161/hv.21703. Epub 2012 Aug 21. PMID: 22906939; PMCID: PMC3656071. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3656071/ Kumar R, Parray HA, Shrivastava T, Sinha S, Luthra K. Phage display antibody libraries: A robust approach for generation of recombinant human monoclonal antibodies. Int J Biol Macromol. 2019 Aug 15;135:907-918. doi: 10.1016/j.ijbiomac.2019.06.006. Epub 2019 Jun 3. PMID: 31170490. https://www.sciencedirect.com/science/article/abs/pii/S0141813019330855?via%3Dihub Ledsgaard L, Kilstrup M, Karatt-Vellatt A, McCafferty J, Laustsen AH. Basics of Antibody Phage Display Technology. Toxins (Basel). 2018 Jun 9;10(6):236. doi: 10.3390/toxins10060236. PMID: 29890762; PMCID: PMC6024766. https://www.mdpi.com/2072-6651/10/6/236 Saw PE, Song EW. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell. 2019 Nov;10(11):787-807. doi: 10.1007/s13238-019-0639-7. Epub 2019 May 28. PMID: 31140150; PMCID: PMC6834755. https://link.springer.com/article/10.1007/s13238-019-0639-7 Sioud M. Phage Display Libraries: From Binders to Targeted Drug Delivery and Human Therapeutics. Mol Biotechnol. 2019 Apr;61(4):286-303. doi: 10.1007/s12033-019-00156-8. PMID: 30729435. https://link.springer.com/article/10.1007/s12033-019-00156-8 Wu CH, Liu IJ, Lu RM, Wu HC. Advancement and applications of peptide phage display technology in biomedical science. J Biomed Sci. 2016 Jan 19;23:8. doi: 10.1186/s12929-016-0223-x. PMID: 26786672; PMCID: PMC4717660. https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-016-0223-x
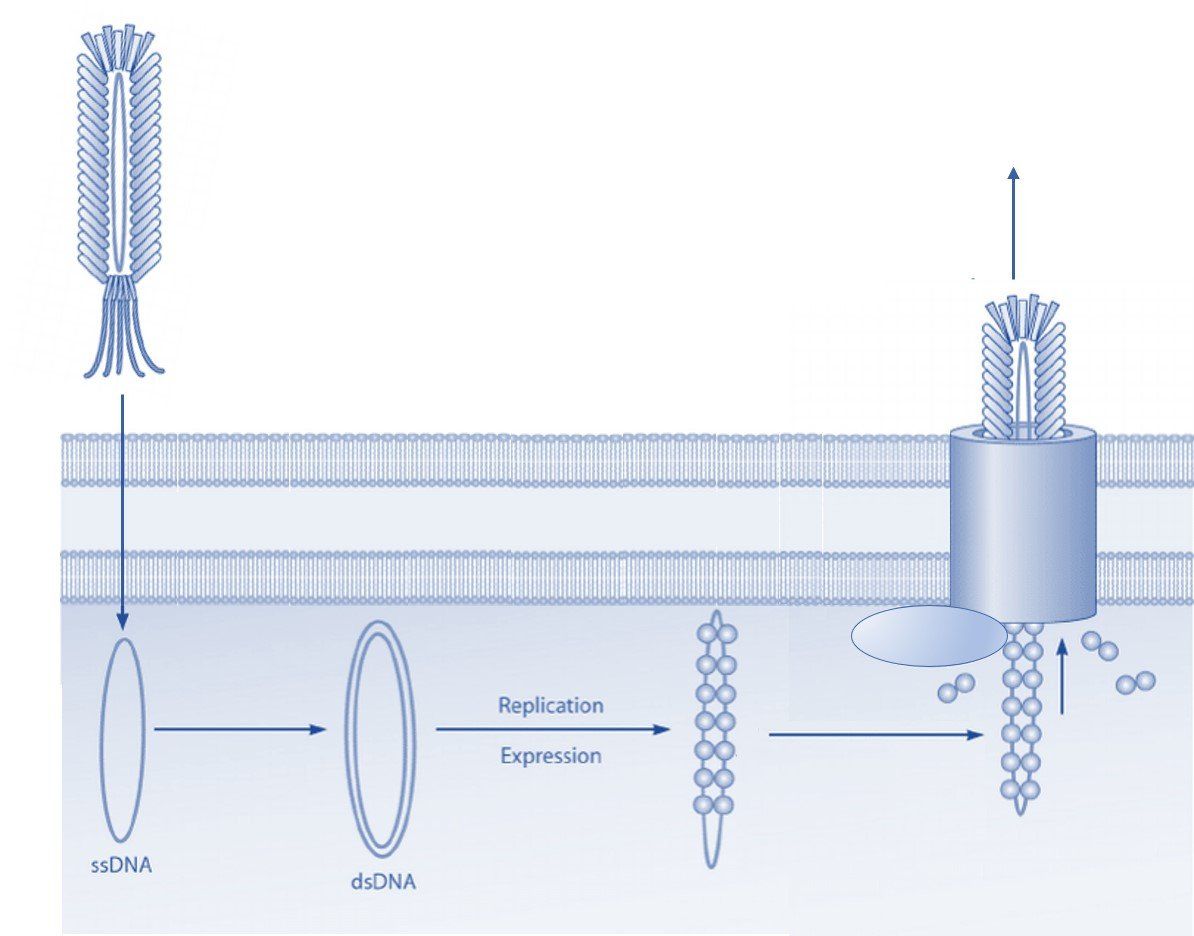
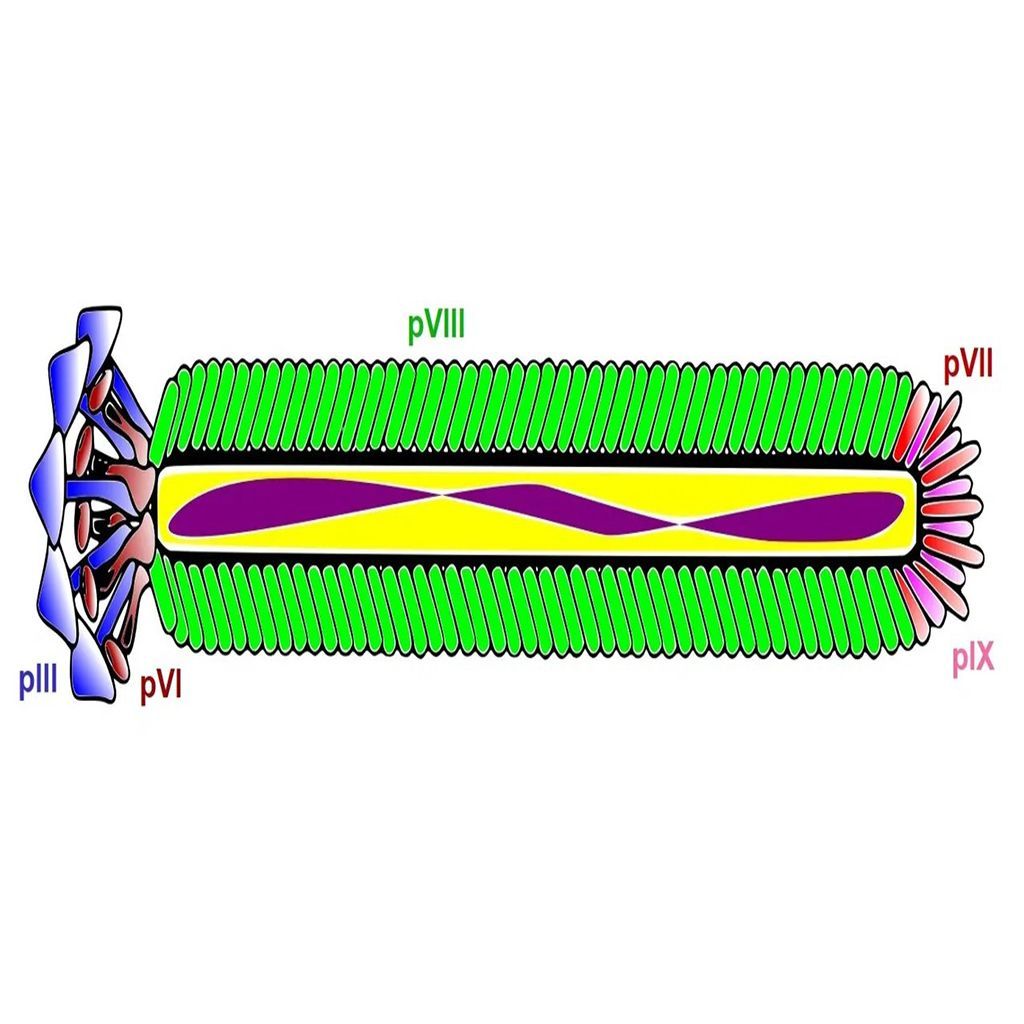
Phage display technology was developed by Dr. George P. Smith in 1985 at the University of Missouri. He genetically modified the filamentous phage genome to display foreign peptide sequences on coat protein III (pIII). Phage display technology was further advanced by creating large random phage libraries that can be used in biopanning for affinity selection of high-specificity peptides and antibodies. Today phage display technology continues to be used in drug discovery for the high-throughput selection of polypeptide ligands including peptides, antibodies, antibody fragments, and nanobodies. Two different types of vectors are typically used for the display of peptides and antibodies on one of the coat proteins (learn more about the different coat proteins used in phage display expression systems here ).