Mette Soendergaard • October 21, 2022
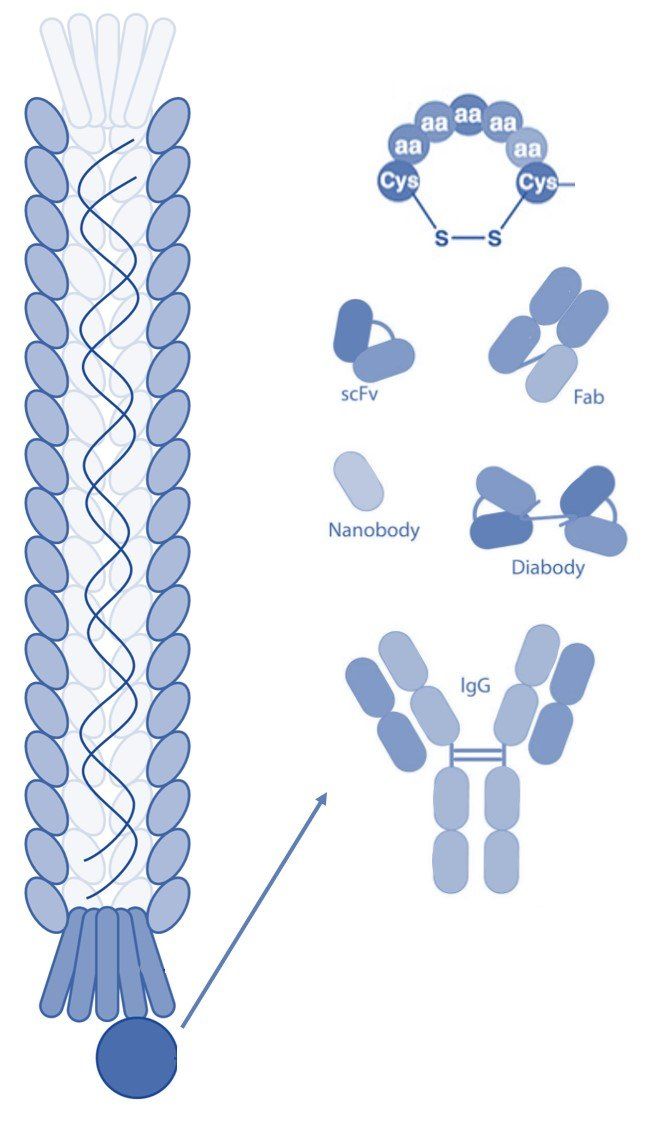
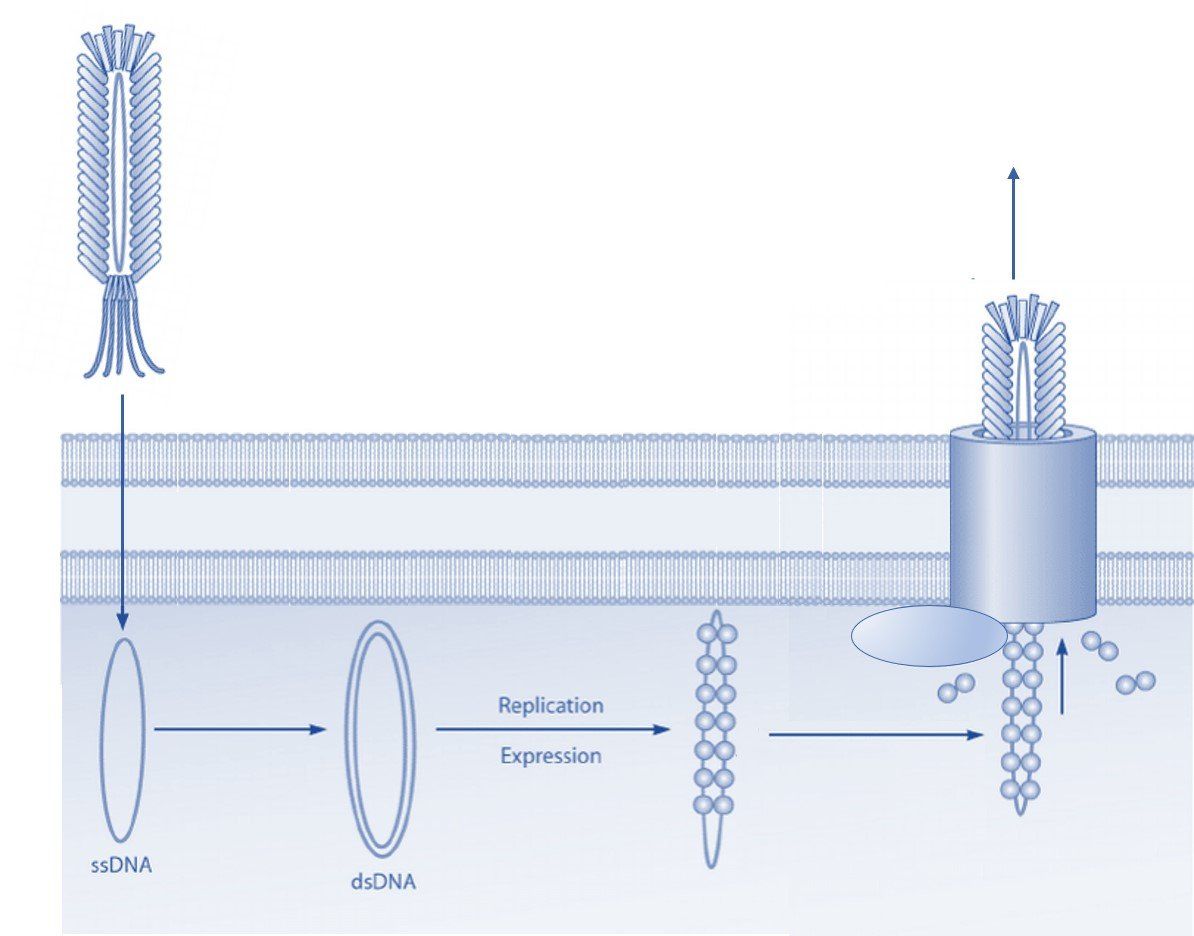
Phage display technology is a laboratory technique that employs genetically modified filamentous bacteriophages to display foreign peptides, antibodies, or other proteins. The phage particles are assembled into phage display libraries, from which researchers can select and identify peptide sequences or antibodies that bind with high affinity and specificity to a target molecule. Phage display technology has been used for many purposes, especially in drug discovery for the development of therapeutic antibodies and peptides.
Biopanning
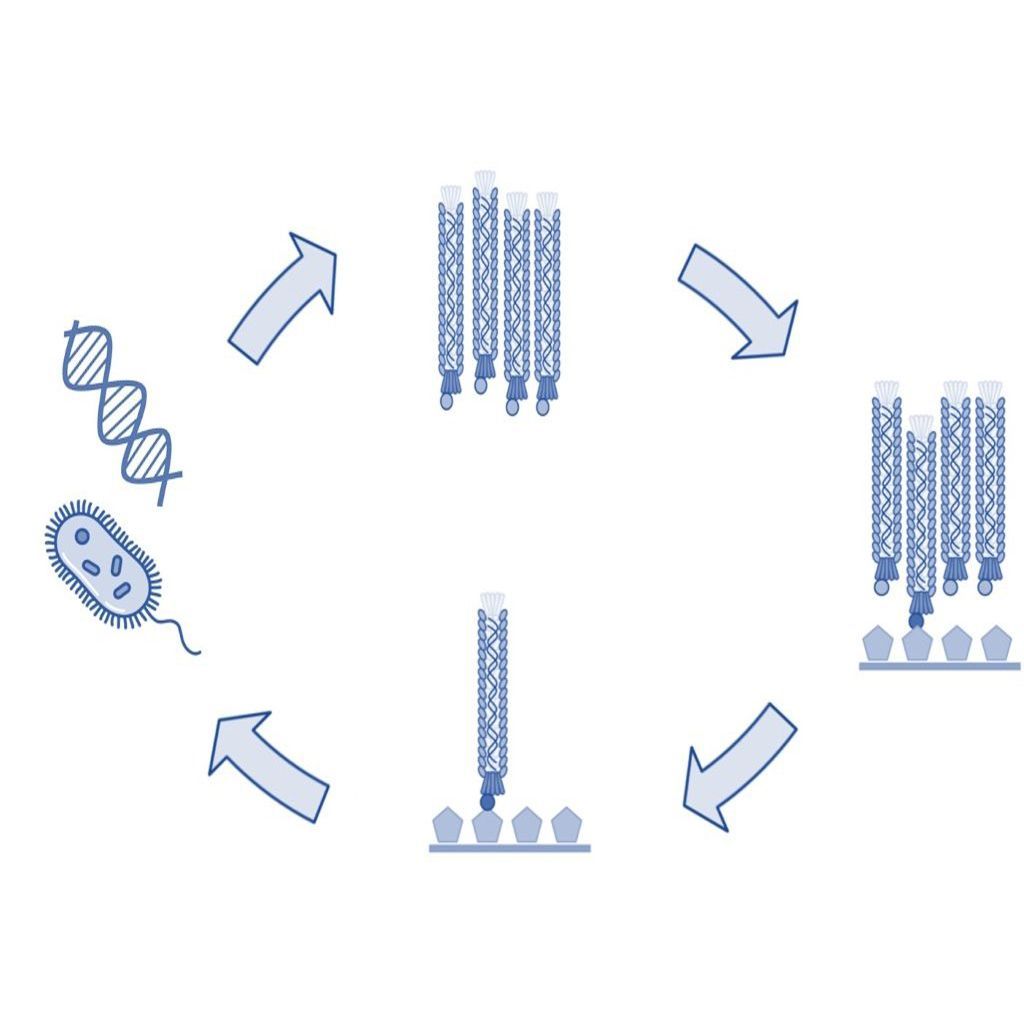
Biopanning of phage libraries is the most common method of selecting and identifying peptides and antibodies that bind to a target antigen. A traditional biopanning procedure includes the incubation of a phage library with the target of interest followed by the collection of bound phages by elution. Each biopanning protocol contains the following key steps, which are described in more detail below.
- Incubation of the phage display library with the target of interest (affinity selection).
- Removal of unbound phage by washing.
- Collection of bound phage by elution.
- Amplification of collected phage.
- Repetition of steps 1-4 for typically a total of four rounds.
- Identification of collected phage by DNA sequencing.
While phage display biopanning is a relatively simple and efficient technique, it requires careful optimization of several parameters in order to achieve the best results. These include the choice of the type of phage library, phage and target molecule concentration, cell type (tumor cells, bacterial cells, etc.), incubation conditions such as temperature and time, as well as the amplification method of collected phage.
Affinity Selection
The first step of a biopanning process is often referred to as affinity selection since this step allows phage-displayed peptides or antibodies with affinity for the target to bind. Often, the target molecule is an immobilized recombinant protein. However, carbohydrates, lipids, and other biomolecules can also be used. Additionally, whole cells, tissues, and organisms (experimental animals, human patients, etc.) have been used with great success in biopanning protocols to identify binding peptides and antibodies with high affinity and specificity.
For affinity selection, it is important to consider the stringency of the selection, which can be modified by changing the concentration of either the phage libraries or the target antigens. For example, to select high-affinity ligands binding competition between the different phage clones may be increased by raising the concentration of the phage library or by lowering the concentration of the target molecule. Such stringent conditions can significantly increase the likelihood of identifying desirable phage clones.
Removal of Unbound Phages
Unbound phages are typically removed by extensive washing using a buffer with a low concentration of a detergent. Weakly bound phages, which are typically undesirable in the phage display selection, can be removed by increasing the concentration of detergent or adding additional washing steps.
Elution of Bound Phages
In standard biopanning procedures, bound phages are collected by elution using detergents, changes in pH, or other methods of disrupting the non-covalent interactions between the phage and target moiety. Using such elution methods generally results in the collection of the vast majority of bound phages. However, phage clones with high binding affinity may not be sufficiently eluted by disruption of the non-covalent interactions. Thus, trypsin digestion has become prevalent and ensures equivalent elution of the bound phages.
Amplification of Phages
After each selection round, eluted phages are usually amplified and used in subsequent selections. Amplification of collected phages can be done in different ways and depends on the type of vector used for the phage library. Bacteriophage vectors such as the fUSE5 library are most often amplified by the transduction of E. coli by the phage particle. Transformation by E. coli electroporation of the bacteriophage vector is typically complicated by the large size of the filamentous phage genome. Thus, this method often results in low transformation rates and valuable clones may be lost in this manner. Phagemid vectors are significantly smaller than bacteriophage vectors and are, therefore, more easily transformed into E. coli cells. However, for the propagation of phage particles, a helper phage is required to ensure the assembly and release of the viral progeny.
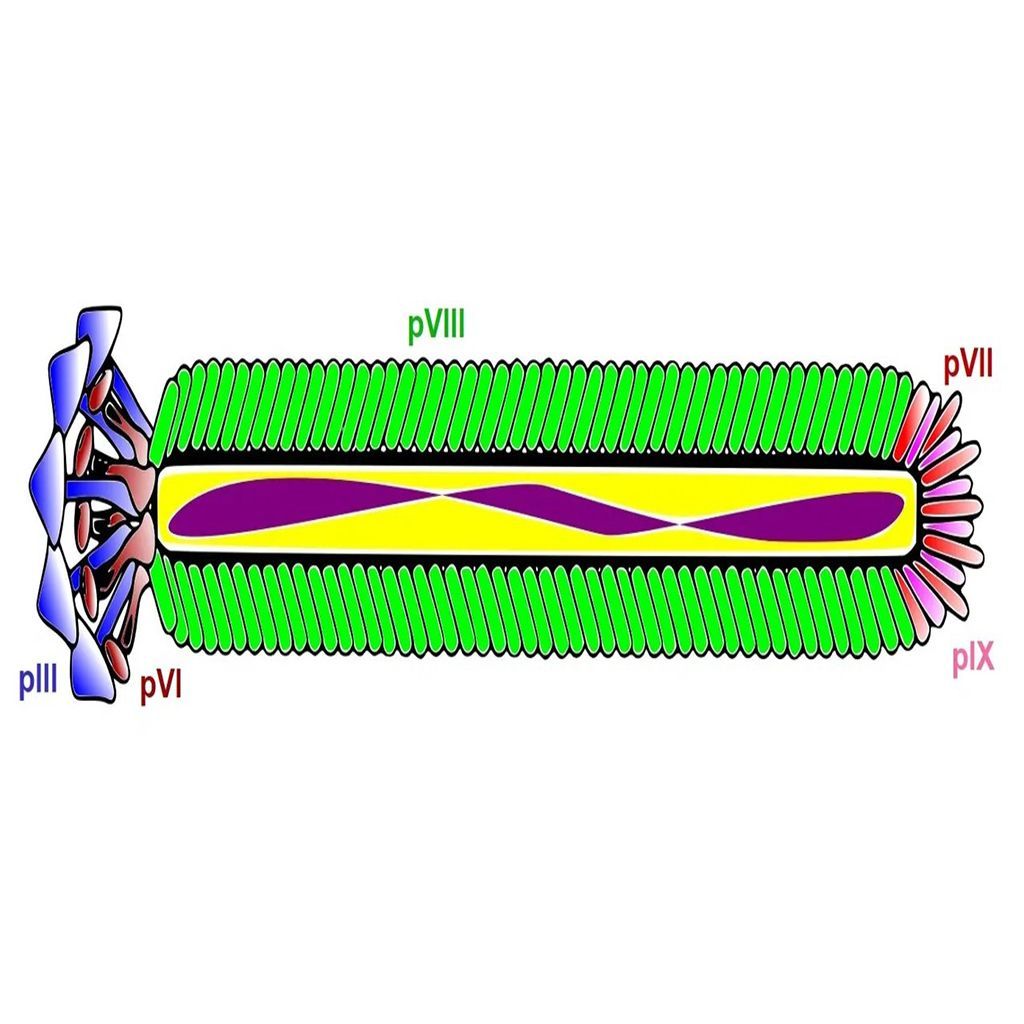
Amplification of selected phage in E. coli is an easy and efficient method. However, the infection of E. coli can be influenced by the phage-displayed peptide or antibody. The infection of E. coli by filamentous bacteriophage is mediated by the interaction between the phage coat protein III (pIII) and the bacterial F pili. The use of the popular pIII-display libraries can thus lead to a change in the infection efficiency and create a selection bias towards certain phage clones. This selection bias may be overcome by using PCR to amplify the foreign oligonucleotide sequences encoding the displayed peptide or antibody. The PCR amplicons can then be inserted back into the bacteriophage vector or phagemid for subsequent transformation and propagation in E. coli.
Repeating the Selection Rounds
Most biopanning procedures include multiple rounds of affinity selection. Typically, four rounds of biopanning against the target antigen are employed to ensure sufficient selection pressure. However, fewer rounds of affinity selection may be used if the protocol includes deep next-generation sequencing of the eluted phages and bioinformatic analysis to sort the identified sequences.
Different types of affinity selections may also be used in biopanning to identify phage clones with specific binding characteristics. For example, phage display selection against a recombinant protein may be combined with a selection round using a cell line that expresses the target protein on the cell surface. Such multi-tier biopanning strategies increase the likelihood of selecting peptides and antibodies that have optimal binding properties both in vitro and in vivo.
Identification of Phage Clones
Eluted phages from affinity selections are identified by DNA sequencing of the foreign inserts that encode the phage-displayed peptides or antibodies. Typically, only the phage clones from the last round of affinity selection are subjected to DNA sequencing. However, it can be an advantage to sequence the phage mixtures after each round of selection. Doing so allows the researcher to follow the efficiency of the phage display procedure since phage particles with binding affinity for the target should become more populous as the selection proceeds.
In addition, it is advantageous to identify unbound phages that are removed from the selection by washing. This step helps identify target-unrelated peptides (TUP) and antibodies that may otherwise be incorrectly identified as potential leads. It may also identify so-called over-growers that exhibit a propagation advantage by infecting E. coli cells at a comparatively higher rate.
Summary
Biopanning is the most common phage display strategy used to select antibodies and peptides with high affinity and specificity for a target molecule. Biopanning is done in several steps that include affinity selection, removal of unbound phages by washing, elution and amplification of bound phages, as well as DNA sequencing to identify selected phage clones. While biopanning can be straightforward, several considerations are important, such as the choice of peptide library or antibody library, the affinity selection conditions, and the extent of DNA sequencing. When taking these considerations into account, biopanning can be a powerful tool for discovering therapeutic antibodies and peptides.
Do you need a custom peptide, antibody, or biopanning strategy for your research?
Cell Origins is the perfect partner for advanced biopanning strategies. Our scientists have years of experience in phage display technology and will work with you to find the best solution for your needs.
With our multi-tier phage display selection process, we can provide you with the most optimal peptides and antibodies for your research with superior binding characteristics and pharmacokinetics. Contact us today to discuss your research needs.
Learn More
Ehrlich GK, Berthold W, Bailon P. Phage display technology. Affinity selection by biopanning. Methods Mol Biol. 2000;147:195-208. doi: 10.1385/1-59259-041-1:195. PMID: 10857097.
https://pubmed.ncbi.nlm.nih.gov/10857097/
Li Y, Liu M, Xie S. Harnessing Phage Display for the Discovery of Peptide-Based Drugs and Monoclonal Antibodies. Curr Med Chem. 2021;28(40):8267-8274. doi: 10.2174/0929867327666201111144353. PMID: 33176631.
https://www.eurekaselect.com/article/111400
Lim CC, Woo PCY, Lim TS. Development of a Phage Display Panning Strategy Utilizing Crude Antigens: Isolation of MERS-CoV Nucleoprotein human antibodies. Sci Rep. 2019 Apr 15;9(1):6088. doi: 10.1038/s41598-019-42628-6. PMID: 30988390; PMCID: PMC6465254.
https://www.nature.com/articles/s41598-019-42628-6
McGuire MJ, Li S, Brown KC. Biopanning of phage displayed peptide libraries for the isolation of cell-specific ligands. Methods Mol Biol. 2009;504:291-321. doi: 10.1007/978-1-60327-569-9_18. PMID: 19159104; PMCID: PMC4053471.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4053471/pdf/nihms326434.pdf
Perera DJ, Scully C. The phage display of peptides and proteins: a practical approach. Curr Protoc Mol Biol. 2014;105:2.44.1-2.44.12. doi: 10.1002/0471142727.mb0244s105. PMID: 2448