Customized Phage Display Libraries
Our custom libraries are made with built-in options for post-selection analysis such as biotinylation, expression of soluble ligands, affinity isolation, and more. This allows you to get the most out of your peptide or antibody phage display library and get the best results. Our experts are here to help you every step of the way, ensuring that you get the most optimal phage display library for your needs.

Leading Experts in Phage Display
At Cell Origins, we specialize in creating custom peptide and antibody phage display libraries to maximize the potential of your selections. Our dedicated team collaborates with you to design and construct a customized library tailored to your unique requirements.
By incorporating selection and tagging strategies directly into the phage display libraries, we facilitate the identification of peptides and monoclonal antibodies with superior affinity, specificity, as well as other essential properties for both
in vitro and
in vivo applications. Moreover, these integrated features enable efficient post-selection analysis, including high-throughput tagging and screening, ensuring rapid and efficient results.
Endless Possibilities with Customized Libraries
Phage display can be used for a variety of applications at different stages in drug development including peptide and antibody discovery, drug candidate screening, and lead optimization. Phage display technology is versatile because almost any type of peptide or antibody library can be screened against any type of target. Additionally, the phage display platform is highly efficient and can be used to screen billions of different peptides or antibodies in a single day. This high throughput capacity makes phage display an attractive technology in drug discovery. However, some important considerations need to be made before choosing which type of library to use for your phage display selections.
Choosing the Right Phage Display Format
The structure of the filamentous phage offers many opportunities for the display of polypeptide-based ligands such as peptides, antibodies, antibody fragments, vhh/nanobody, and more. While the expression of foreign ligands on coat proteins pIII and pVIII is the most common, the utilization of each coat protein presents its own advantages.
pIII
Coat protein III (pIII) is located at one end of the phage virion and acts as the f-pili-binding protein during infection of E. coli. This process is crucial for the successful infection of E. coli and ultimately, for their propagation. Despite its key involvement in bacterial infection, pIII can successfully be used for display without significantly affecting the phage lifecycle. This property ensures the propagation of each clone in the phage display library, maintaining the sequence diversity of the library. For this reason, pIII is the most commonly used display format in phage display technology.
pVIII
Coat protein VIII (pVIII), also known as the major coat protein, is expressed in over 2000 copies, providing ample opportunities to display multiple copies of a peptide. However, not all 2000 copies are typically used for phage display. Rather, only ~10% of the pVIII copies are used for expression. The display of large proteins such as full-length monoclonal antibodies and antibody fragments is challenged due to the impairment of virion release from E. coli during the phage lifecycle. Nevertheless, certain types of peptide selection strategies, such as those that require the binding of polyvalent antigens.
pVI and pVII
Coat protein VI (pVI) is adjacent to pIII. While comparatively less common, pVI display formats have proven useful for expressing larger proteins at both the N- and C-termini. Notably, this type of display has been used to create cDNA libraries and investigate protein-molecule interactions. Coat protein VII (pVII) is located at the opposite end of the virion. Display using pVII has been gaining popularity lately, particularly for expressing antibodies and antibody fragments. Moreover, using pVII display in conjunction with other display techniques can streamline tagging and post-screening analysis.
pIX
Recent advancements have significantly enhanced the display on coat protein IX (pIX) through the utilization of diverse phagemid systems and signal sequences. However, when compared to other systems, particularly those employing pIII, the expression levels of foreign polypeptides on pIX tend to be lower. Nevertheless, this lower expression level increases the likelihood of monovalent display, thereby facilitating the selection of high-affinity ligands. This progress underscores the importance of optimizing the display system for continuously improved results.
Choosing an Optimal Expression System
Dr. George P. Smith pioneered the development of phage display technology in 1985 at the University of Missouri. His groundbreaking work involved genetically modifying the filamentous phage ssDNA genome to express foreign peptide sequences on coat protein III (pIII). Expanding on this innovation, large random phage libraries were created, leading to the development of biopanning for the selective identification of highly specific peptides and antibodies. Today, two distinct types of vectors are commonly utilized for the display of foreign polypeptide-based ligands.
Bacteriophage Vectors
Filamentous bacteriophage vectors (Fd and M13) encode a modified version of the complete phage genome. Alongside the wild-type genes, most bacteriophage vectors have an antibiotic selection gene. Additionally, they carry a recombinant hybrid gene that encodes the foreign peptide or antibody fused to the coat protein utilized for display. Expression of both the wild type and hybrid genes facilitates a lower level of foreign polypeptide display, which bears the advantage of enabling the selection of high-affinity polypeptide. However, bacteriophage vectors are not be ideal for displaying antibodies or antibody fragments due to their large size, which impedes E. coli infection and complicates transformation. Nevertheless, bacteriophage vectors have proven successful in peptide phage display. A notable advantage is that these vectors do not necessitate the use of helper phages for phage particle propagation, as their genome encompasses all the required replication genes.
Phagemids
Phagemids are vectors based on plasmids that contain phage and bacterial origins of replication, along with an antibiotic selection gene. Moreover, a phagemid vector typically encodes a coat protein fused to a foreign DNA sequence. This fusion is used for the display of monoclonal full-length antibodies, antibody fragments, nanobodies, or peptides. Phagemid vectors do not rely on helper phage proteins to replicate in
E. coli. However, helper phages are indispensable for producing phage clones, as they carry essential genes required for the assembly and release of the phage particles. The small size of phagemids facilitates hassle-free transformation in E. coli. The efficiency of
E. coli transformation directly impacts the diversity of the phage display library, making it a crucial consideration when selecting a suitable vector. Additionally, the small size of phagemid vectors allows for easy manipulation using recombinant DNA technology.
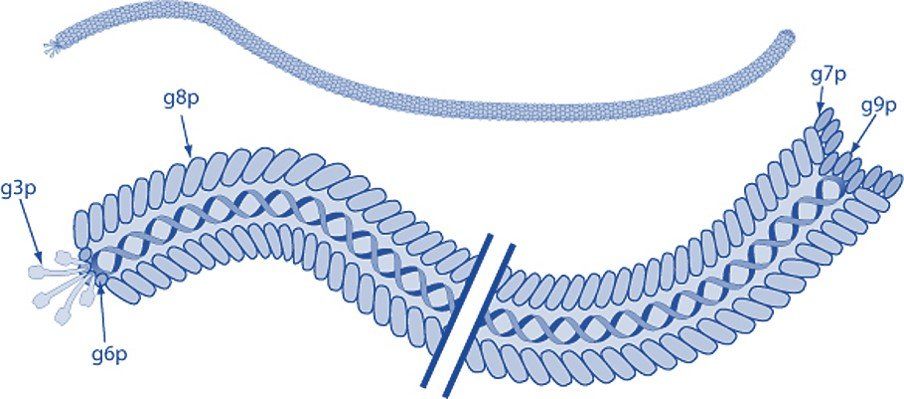
Filamentous bacteriophage express five coat proteins that can each be used to display foreign polypeptides and employed in phage display technology. Coat protein VIII (pVIII) is the major coat protein and is expressed in >2000 copies along the virion. Coat proteins III and VI are expressed at one tip of the bacteriophage, while pVII, and p IX are located at the opposite end.
Modified from ViralZone, SIB Swiss Institute of Bioinformatics. Permission to use ViralZone graphics in Wikimedia Commons kindly granted by SIB (legal(at)sib.swiss), 2021-02-01. -
https://viralzone.expasy.org/resources/Inoviridae%5Fvirion.jpg on https://viralzone.expasy.org/113, CC BY-SA 4.0,
https://commons.wikimedia.org/w/index.php?curid=99827306
Phage Display Peptide & Antibody Libraries
Peptide phage display technology is an efficient technology with a broad range of applications in research and drug discovery. Peptides can serve as drugs to induce therapeutic effects or transport cargo to target disease biomarkers. The compact nature of peptides presents numerous benefits, such as their ability to penetrate tissues and be easily eliminated from the body. These advantages are particularly crucial in cancer radiotherapy, where precise delivery to target cells and swift clearance are vital to prevent damage to non-targeted tissues. Antibody phage display libraries are invaluable tools in the field of drug discovery and scientific research. These versatile libraries facilitate the screening of antibodies and peptides, identifying those with exceptional binding affinities and specificities. With the ability to target a diverse array of antigens, including proteins, peptides, small molecules, and even whole cells and tissues, these libraries offer an enormous potential for advancing biomedical knowledge.

The Success of Linear Peptides
Phage display technology enables the selection of both linear and cyclic peptide sequences, each offering distinct advantages and disadvantages. Linear phage display peptide libraries, which have been available for decades, are generally more affordable compared to their cyclic counterparts. Moreover, the synthesis of soluble linear peptides is often easier and more cost-effective, thereby providing a convenient option for researchers. However, their limited bioavailability due to rapid degradation in the body can hamper their efficacy as drugs unless they undergo substantial modifications.

Harness the Power of Cyclic Peptides
Over the past decade, there have been notable advancements in the development of novel cyclic peptide libraries using phage display. These libraries are constructed by inducing disulfide bond formation among cysteine residues or through the utilization of scaffolding systems. Cyclic peptides possess exceptional stability owing to their non-natural structure, which also provides them with a rigid framework resulting in improved target binding affinity and specificity. Consequently, they prove to be an excellent choice for applications requiring high potency or selectivity, such as cancer treatments or addressing neurological disorders like Alzheimer's disease. Despite their benefits, it is worth noting that the complex synthesis process involved in creating cyclic peptides can be consuming both time-wise and financially.
Monoclonal Antibodies
Full-length monoclonal antibodies have emerged as an important category of biopharmaceuticals, holding a significant position in the market. Interestingly, this class accounts for six out of the top ten best-selling drugs. Leveraging large Ig gene repertoires, fully human antibodies can be assembled within phage display libraries to carefully select antibodies with desirable traits. The utilization of phage display antibody libraries presents distinct advantages over traditional hybridoma technology, which traditionally have produced murine-derived monoclonal antibodies that often demonstrate limited therapeutic efficacy due to rapid clearance and allergic reactions.
Expansive Options
The development of smaller recombinant antibody fragments, such as variable domain (Fv), single-chain variable domain (scFv), diabodies (bivalent scFvs), fragment antigen binding (Fab), and heavy-domain camelid, alpaca, llama, or shark antibody VHH fragments (nanobodies) have helped further advance antibody phage display. These antibody fragments are more amenable to expression than full-length antibodies that require the assembly of four polypeptide chains and extensive disulfide bond formation.
Let's Get Started!
At Cell Origins, we closely collaborate with you to create a personalized peptide or antibody phage display library tailored to your unique objectives. Our dedicated team of experts incorporates state-of-the-art selection and tagging strategies into the library to enable the identification of ligands with exceptional binding kinetics and pharmacokinetics. This streamlines the process of screening, ensuring accuracy and reliability of your phage display selections. Throughout the development of your customized phage display library, our team provides comprehensive support and protocols. Whether you prefer to schedule a meeting or reach out to us directly, we are eager to learn more about your research needs. Trust us to develop a custom phage display library that meets your goals.
-
Published Work
Axiak-Bechtel, S. M., Leach, S. B., Scholten, D. G., Newton-Northup, J. R., Johnson, B. J., Durham, H. E., Gruber, K. A., Callahan, M. F. (2021). Pharmacokinetics and safety of TCMCB07, a melanocortin-4 antagonist peptide in dogs. Pharmacology Research & Perspectives. Vol. 9(3)
Asar, M., Franco, A., Soendergaard, M. (2020). Phage Display Selection, Identification, and Characterization of Novel Pancreatic Cancer Targeting Peptides. Biomolecules. 10(5), 714
Asar, M., Gunby, T., Franco, A., Woodson, C., Soendergaard, Mette. (2020). Identification of an Indiscriminate Peptide by Phage Display Technology. The FASEB Journal 34:1_supplement, 1-1
-
More
Asar, M., Newton-Northup, J., Deutscher, S., Soendergaard, M. (2019). Ovarian Cancer Targeting Phage for In Vivo Near-Infrared Optical Imaging. Diagnostics, 9, 183.
Newton-Northup, J.R., and Deutscher, S.L. (2017). Bacteriophage for the Development of Novel Tumor-Targeting Agents with Specific Pharmacokinetics and Imaging Applications. Methods in Molecular Biology in Biosensors and Biodetection.
Newton-Northup, J.R., and Deutscher, S.L.. (2016). Cytotoxic Tumor-Targeting Peptides From In Vivo Phage Display. Combinatorial Chemistry & High Throughput Screening. Vol. 19(5): 370–377
García, M. F., Zhang, X., Shah, M., Newton-Northup, J., Cabral, P., Cerecetto, H., and Quinn, T. (2016). 99mTc-bioorthogonal click chemistry reagent for in vivo pretargeted imaging. Bioorganic & Medicinal Chemistry. Vol. 24(6): 1209–1215
Newton-Northup, J.R., Dickerson, M.T., Kumar, S.R., Smith, G.P., Quinn, T.P., And Deutscher, S.L. (2014) In vivo bacteriophage peptide display to tailor pharmacokinetics of biological nanoparticles. Molecular Imaging and Biology. 16(6), 854-864.
Soendergaard M., Newton-Northup J. R., Deutscher S. L. (2014): In Vivo Phage Display Selection of an Ovarian Cancer Targeting Peptide for SPECT/CT Imaging. American Journal of Nuclear Medicine and Molecular Imaging. 4(6): 561–570.
Soendergaard, M., Newton-Northup, J.R., and Deutscher, S.L. (2014) In vitro high throughput phage display selection of ovarian cancer avid phage clones for near-infrared optical imaging. Combinatorial Chemistry and High Throughput Screening. 17(10).
Newton-Northup, J.R., and Deutscher, S.L. (2013). Contending With Target Unrelated Peptides from Phage Display. Journal of Molecular Imaging & Dynamics. Vol. 2(2):
Newton-Northup, J. R., Dickerson, M. T., Ma, L., Besch-Williford, C. L., Deutscher, S. L. (2013). Inhibition of metastatic tumor formation in vivo by a bacteriophage display-derived galectin-3 targeting peptide. Clinical & Experimental Metastasis. Vol. 30:119–132
Newton-Northup, J.R., and Deutscher, S.L. (2012) Contending with target unrelated peptides from phage display. Journal of Molecular Imaging and Dynamics. 2(2).
Soendergaard, M., Newton-Northup, J.R., Palmier, M.O., and Deutscher, S.L. (2011) Peptide phage display for discovery of novel biomarkers for imaging and therapy of cell subpopulations in ovarian cancer. Journal of Molecular Biomarkers and Diagnosis, S:2.
Newton-Northup, J.R., Figueroa, S.D., and Deutscher, S.L. (2010). Streamlined in vivo selection and screening of human prostate carcinoma avid phage particles for development of peptide based in vivo tumor imaging agents. Combinatorial chemistry & high throughput screening. 14(1):9-21
Deutscher, S. L., Dickerson, M., Gui, G., Newton, J., Holm, J. E., Vogeltanz-Holm, N., Kliethermes, B., Hewett, J. E., Kumar, S. R., Quinn, T. P., Sauter, E. R. (2010). Carbohydrate antigens in nipple aspirate fluid predict the presence of atypia and cancer in women requiring diagnostic breast biopsy. BMC Cancer. Vol. 10(519)
Newton-Northup, J.R., Figueroa, S.D., Quinn, T. P., and Deutscher, S.L. (2009). Bifunctional phage-based pretargeted imaging of human prostate carcinoma. Nuclear Medicine and Biology. Vol. 36(7):789-800
Jin, X., Newton, J. R., Montgomery, S., Smith, G. P. (2009). A generalized kinetic model for amine modification of proteins with application to phage display. Biotechniques. Vol. 46(3):175-182
Newton-Northup, J.R., and Deutscher, S.L. (2009). In vivo bacteriophage display for the discovery of novel peptide-based tumor-targeting agents. Methods in Molecular Biology. Vol. 504:275-90.
Newton-Northup, J.R., and Deutscher, S.L. (2008). Peptide Phage Display. Handbook of Experimental Pharmacology: Molecular Imaging I. Vol. 185 Pt 2:145-163.
Newton, J. R., Miao, Yubin., Deutscher, S. L., and Quinn, T. P. (2007). Melanoma Imaging with Pretargeted Bivalent Bacteriophage. Journal of Nuclear Medicine. Vol. 48(3):429-436
Newton, J. R., Kelly, K. A., Mahmood, U., Weissleder, R., and Deutscher, S. L. (2006) In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia. 8(9), 772-780.
Contact Us Any Time
Contact Us
We will get back to you as soon as possible
Please try again later
